Last updated on Nov. 16, 2025 at 03:44 p.m.
Researchers at the University created an artificial intelligence model named EZSpecificity to test how well certain enzymes and substrates bind to each other.
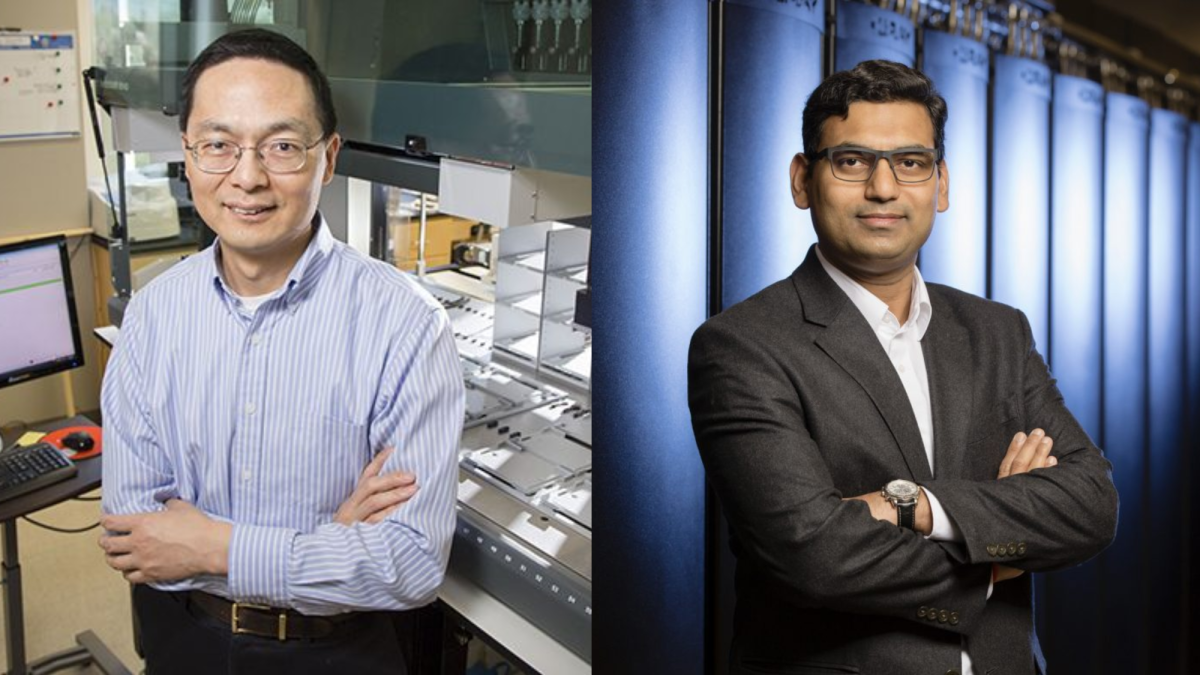
Huimin Zhao and Diwakar Shukla, professors in Engineering, worked to develop the EZSpecificity model — an AI system that matches chemicals to specific enzymes. They say the tool can be used to advance drug development and synthetic biology.
Zhao’s research focus involves using synthetic biology and machine learning to create novel chemical reaction mechanisms. Shukla’s research centers on explaining complex biological processes using physics-based models and techniques.
Machine learning is a form of AI that can use given data and “learns” from it to make predictions about the data. Synthetic biology is redesigning molecules so they can produce new products, such as medicine.
Get The Daily Illini in your inbox!
What is EZSpecificity, and how does it work?
Enzymes are proteins that help accelerate chemical reactions. They act upon chemical compounds called substrates to facilitate these reactions. Many of the day-to-day biological functions the human body performs are reactions catalyzed, or accelerated, by enzymes.
The EZSpecificity model can be used to find pathways to develop chemicals for new drugs or find the optimal substrate for an enzyme, according to Zhao. The model uses AI to predict which chemicals can be substrates for which enzyme.
Zhao said it’s often unclear which substrate works with a particular enzyme. EZSpecificity, Zhao hopes, will help the researchers figure it out.
“Inside the cell, sometimes you don’t know which enzymes really catalyze which reactions — and then we can also help to figure out the metabolism,” Zhao said.
Understanding certain enzyme-substrate relationships can help advance drug discovery and other biological research. Zhao further said the technology could be applied to a broad range of fields, including enzyme engineering, synthetic biology and biocatalysis.
Shukla said the process requires very extensive experimentation to establish which chemicals could be modified, as making the enzymes and chemicals and testing them is both tedious and precise. He added that a highly efficient AI tool can be incredibly valuable to both researchers and the public.
“If you want to make a particular chemical or a product or some type of drug, you can predict what kind of enzyme will make that product and what kind of reactants are needed to make it,” Shukla said.
How was it trained?
The EZSpecificity model was trained using two datasets: PDBind+ and ESIBank. These datasets combined lots of information about enzymes, substrates and their reactions for the model to train on and be more accurate.
“Where we try to predict using molecular docking and simulations, we tried to predict which substrate binds to which enzyme,” Shukla said. “Then, we created this huge database of enzyme substrate pairs that was purely computational.”
This large-scale computational modeling laid the groundwork for understanding the molecular basis of enzyme-substrate interactions.
“And then (the EZSpecificity model) also understood what kind of interactions are happening because once you have a physical model, you can actually understand the interactions that are allowing this substrate to bind to the enzyme,” Shukla said. “This computational dataset was much bigger than the experimental data set; when you train on both of them together, you get a much more accurate model.”
Dual-input algorithm guides the model
The model uses an algorithm called cross-attention, which operates on two different input sequences: a source sequence and a target sequence. The algorithm is typically used within the decoder layers of a large-language model, where the source sequence is the context and the target sequence is the sequence being generated.
In the case of EZSpecificity, this describes the interactions between specific substrate chemical groups and enzyme amino acid residues. Given an enzyme-substrate complex — the source sequence — the model predicts the specific interactions between the two.
“(It) predicts what kind of amino acids and chemical group interactions would happen if you give it a structure of an enzyme-substrate complex,” Shukla said.
Zhao and Shukla’s research paper states that the EZSpecificity model computed a 91.7% accuracy in identifying the single potential reactive substrate when validated by experiments. The result was significantly higher than the 58.3% accuracy rating by another AI model, ESP.
Shukla, Zhao seek to share the tool
According to Shukla, the pair is currently developing plans for a website that will be open-source and available to the public.
“Our hope is that this website (will) have a lot of functionalities in the future,” Shukla said. “You can come to our website and just use it open source, no restrictions — but we do have a patent on it.”
Currently, Zhao says he’s implementing EXSpecificity at the Molecule Maker Lab Institute at the University.
Future directions
One of the researchers’ main concerns is that the model’s accuracy is not high enough.
“In the paper, we show the halogenase we use as an example,” Zhao said. “The accuracy is pretty good, more than 95%, 90%, but for other enzymes, the accuracy is low, and we do need to get more data to train the model so that it can be generally applicable with higher accuracy.”
Shukla said that more information should be added about the characteristics of the proposed reactions the model predicts. More specifically, the model does not include energetic information of the reaction, like the Gibbs free energy — the total energy available to drive a reaction.
“Right now what we’re saying is, ‘Oh, this is a substrate,’ or, ‘not a substrate,’ but if it can start predicting the kinetic parameters or the rates of transformation of chemicals, and other similar quantitative measures … that is the next step,” Shukla said. “If you can integrate that type of experimental and computational information into the tool … a highly accurate model for that would be very useful.”